Genomic puzzle of rumen microbes further refined
Ordered genomic map of entire rumen microbiome created to more easily identify individual microbes and their functions.
August 29, 2019
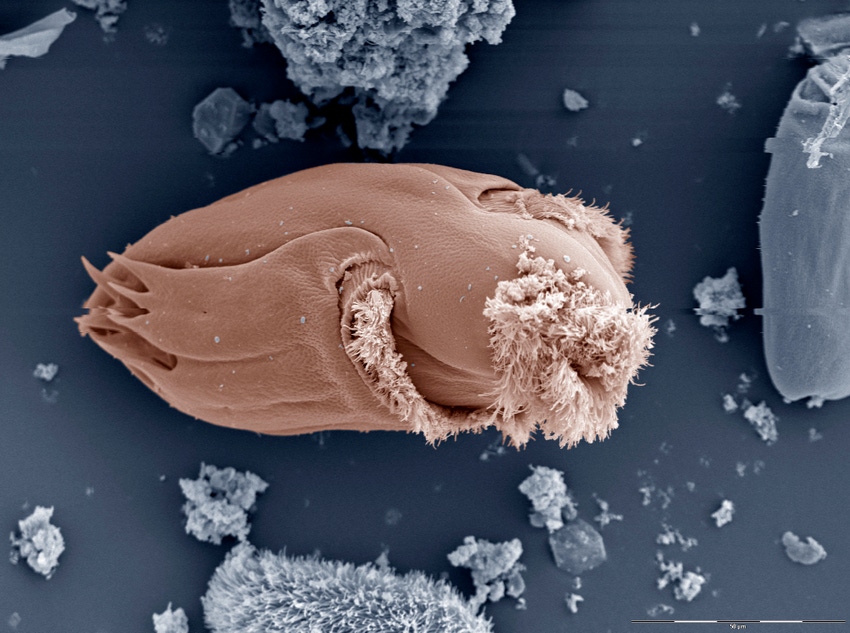
Using high-tech tools, scientists with the U.S. Department of Agriculture's Agricultural Research Service (ARS) and their cooperators have taken a deep dive into the microbial milieu of the cow's rumen.
Ultimately, such efforts could lead to new ways of ensuring the health and well-being of cows as well as improving their production of milk, meat and other products, noted Derek Bickhart, a research microbiologist with the ARS U.S. Dairy Forage Research Center in Madison, Wis.
"Compositionally, the rumen is unique because it contains organisms from all three kingdoms of life: bacteria, archaea and eukaryotes. All of these organisms work in tandem to consume plant matter, and the byproducts of their digestion are used by the cows to produce meat and milk," explained Bickhart, who is at the ARS center's Cell Wall Biology & Utilization Research Unit.
Bickhart estimated that there are more than 30,000 species of these rumen dwellers, yet fewer than a couple thousand have been adequately characterized. That has left a gap in researchers' understanding of what role each species plays as part of the rumen microbiome, ARS said.
Researchers also want to learn what the absence or even the proliferation of a microbe can mean to the cow's well-being and performance — from withstanding costly diseases like mastitis to producing important milkfat.
Now, Bickhart and colleagues are getting a better handle on the situation. Using the latest advances in sequencing equipment, they've devised a way to read the chemical coding of DNA fragments from the microbes and assemble the pieces into whole genomes using special algorithms, ARS said. From this, they created an ordered genomic map of the entire rumen microbiome that will make it easier to identify individual members and their functions.
In the past, these sequence reads were limited to chemical units of measure known as nucleotide base pairs that fell in the 100-500 base pair range. With the new method, the researchers obtained reads of 9,000-30,000-plus base pairs — an increase resulting from the team's use of a long-read commercial sequencer (PacBio Sequel) and the new Proximeta Hi-C method.
Full details appear in the August 2019 issue of Genome Biology.
Among other benefits, longer DNA reads make it easier to identify genes that can reveal what a particular microbe does or how it differs from other closely related species, ARS noted.
"While we haven't completely mapped the genomes of all rumen microorganisms in our study, we've come closer than those in the past and also identified new biological facts about the community that we didn't know before," Bickhart said.
His collaborators and co-authors on the Genome Biology paper include scientists from three other ARS locations, five universities in the U.S. and abroad, two biotech firms and the National Human Genome Research Institute in Bethesda, Md.
Use of the team's specialized DNA extraction and sequencing procedures also revealed new insights about another denizen of the cow rumen: namely, bacteriophages, which are viruses that highjack the genomes of bacteria to replicate themselves. In all, the team identified 188 bacteria/phage interactions, including host preferences, life cycle and impacts on the microbe's metabolic activity.
"On the direct application side, knowledge of the rumen microbial community will be critical to estimating the feed efficiency of cattle and identifying microbes that contribute directly to milk fat production," Bickhart said.
Separately, a group of researchers in the U.K. recently published a compendium of rumen microbes.
Source: Agricultural Research Service, which is solely responsible for the information provided and is wholly owned by the source. Informa Business Media and all its subsidiaries are not responsible for any of the content contained in this information asset.
You May Also Like



