New model of bacterial DNA repeats may lead to vaccine development.
March 12, 2019
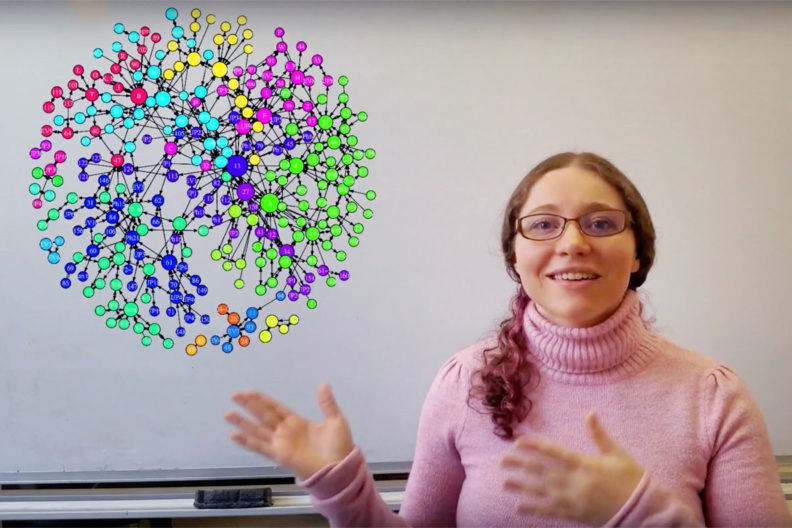
A concise network model developed by Washington State University researchers could help advance the understanding of disease-causing bacteria and guide vaccine development, such as for anaplasmosis in cattle.
Assefaw Gebremedhin, assistant professor in the Washington State School of Electrical Engineering & Computer Science, led a team of researchers in creating the network model to represent similarities between short DNA sequences, according to an announcement.
The team, which included veterinary microbiology and pathology professor Kelly Brayton and computer science graduate student Helen Catanese, used their network model to study the DNA of Anaplasma marginale, a widely distributed tick-borne pathogen that can cause the fatal blood disorder anaplasmosis in cattle.
A. marginale, like many other bacterial pathogens, have a huge variety of strains characterized by their surface protein variation, the researchers said. The team first used short, repeating sequences of DNA, called repeats, from one of these varying surface proteins to characterize strains and build the network. The researchers then used the network to understand the heredity and geographic distribution of the bacterial strains.
They created a visualization of their network, in addition to a graphical representation of the geographic distribution of those same repeats on a map. By comparing this to their network structure, they discovered a previously unknown relationship between how “central†a repeat is in the network and how geographically dispersed it is in the real world, the researchers said.
More “central†repeats — meaning repeats that lie in the pathways of most connections between other repeats in the network — were also found to be more widely distributed in A. marginale strains around the world, ranging from North and South America all the way to Asia.
A paper detailing their work appeared in the journal BMC Bioinformatics.
If the repeats that are structurally important in the network are also the most widely distributed geographically, they could play a role in how easily the pathogen can be transmitted from one animal to another, the researchers said.
“This intriguing finding suggests that these sequences may be functionally important for the organism’s ability to adapt,†Gebremedhin said.
The work can spur future research on why certain repeat sequences became so geographically prevalent and help in targeting vaccine development efforts, he added.
The same modeling and analysis technique can also be extended to other pathogen species that have similar repeating DNA sequences.
Network models have been used to represent similarities among DNA sequence elements in the past, but the new model developed by the Washington State team is sparser and more efficient to construct than existing network models, without losing key structural components, according to the announcement. It maintains its properties even when the sequence data have a large percentage missing or have unwanted disturbances.
Source: Washington State University, which is solely responsible for the information provided and is wholly owned by the source. Informa Business Media and all its subsidiaries are not responsible for any of the content contained in this information asset.
You May Also Like

.png?width=300&auto=webp&quality=80&disable=upscale)

